GMS-Artic
A nextflow pipeline with a GMS touch for running the ARTIC network’s fieldbioinformatics tools
Jyotirmoy Das
Sofia Stamouli
Henning Onsbring
Tanja Normark
Isak Sylvin
Why it requires?
- analyse whole genome sequence data from SARS-CoV-2.
- requires pangolin typing information.
- requires a semi/automated workflow (NextFlow).
- development of production level, clinical data analysis tool
- container-packed (Conda, Docker, Singularity) reproducible package
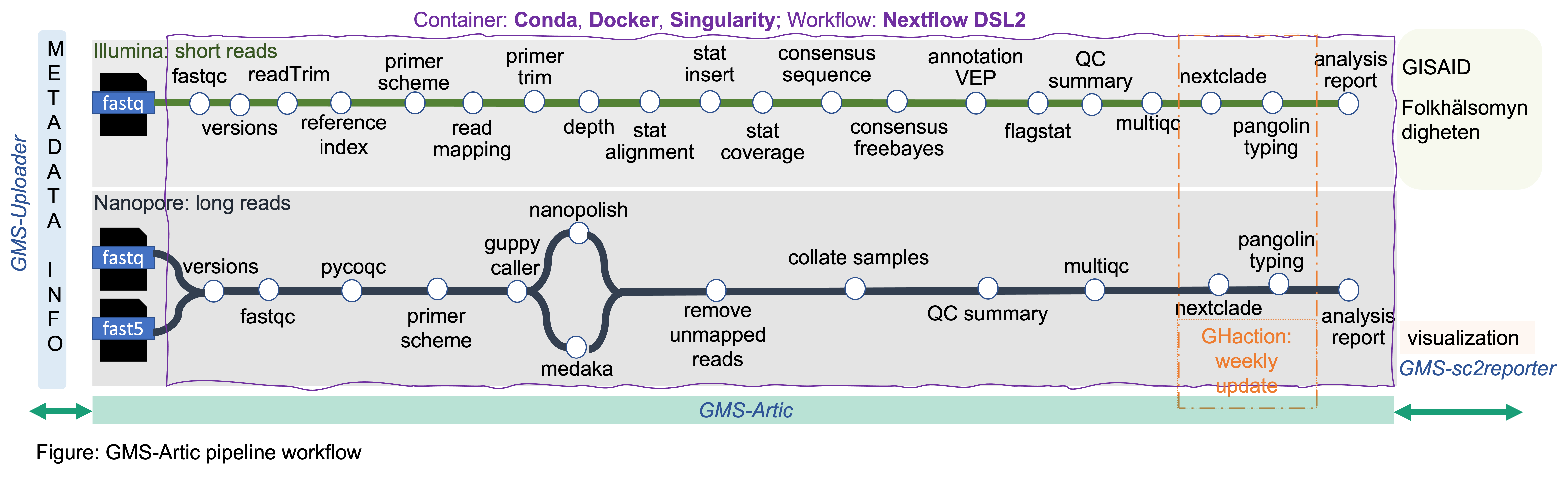
What it does?
Architecture of GMS-Artic
GitHub page

Major updates-v2.0.0
- Docker container separated for Pangolin typing
- Illumina container: gms-artic-illumina
- Nanopore container: gms-artic-nanopore
- Pangolin container: gms-artic-pangolin - bi-weekly updated with GitHub action
- Added separate package version files for each workflow
- versions: for Illumina and Nanopore
- pangoversion: for pangolin typing
- extra features for Illumina, flagstat, depth, VEP annotation
- Illumina results works for sc2reporter visualization
- additional QC features for Nanopore: fastqc, multiqc, pycoQC
How to run?
nextflow run main.nf -profile singularity \ --illumina \ --prefix "test_illumina" \ --directory .github/data/fastqs/ \ --outdir illumina_testnextflow run main.nf -profile singularity \ --illumina \ --prefix "test_illumina" \ --directory .github/data/fastqs/ \ --outdir illumina_testnextflow run main.nf -profile singularity \ --illumina \ --prefix "test_illumina" \ --directory .github/data/fastqs/ \ --outdir illumina_testnextflow run main.nf -profile singularity \ --illumina \ --prefix "test_illumina" \ --directory .github/data/fastqs/ \ --outdir illumina_testnextflow run main.nf -profile singularity \ --illumina \ --prefix "test_illumina" \ --directory .github/data/fastqs/ \ --outdir illumina_testnextflow run main.nf -profile singularity \ --illumina \ --prefix "test_illumina" \ --directory .github/data/fastqs/ \ --outdir illumina_testnextflow run main.nf -profile singularity \ --illumina \ --prefix "test_illumina" \ --directory .github/data/fastqs/ \ --outdir illumina_test
nextflow run main.nf \ -profile singularity \ --nanopolish \ --prefix "test_nanopore_nanopolish" \ --basecalled_fastq /fastq_pass/ \ --fast5_pass /fast5_pass/ \ --sequencing_summary sequencing_summary_FAK72834_298b7829.txt \ --outdir nanopore_nanopolishnextflow run main.nf \ -profile singularity \ --nanopolish \ --prefix "test_nanopore_nanopolish" \ --basecalled_fastq /fastq_pass/ \ --fast5_pass /fast5_pass/ \ --sequencing_summary sequencing_summary_FAK72834_298b7829.txt \ --outdir nanopore_nanopolishnextflow run main.nf \ -profile singularity \ --nanopolish \ --prefix "test_nanopore_nanopolish" \ --basecalled_fastq /fastq_pass/ \ --fast5_pass /fast5_pass/ \ --sequencing_summary sequencing_summary_FAK72834_298b7829.txt \ --outdir nanopore_nanopolishnextflow run main.nf \ -profile singularity \ --nanopolish \ --prefix "test_nanopore_nanopolish" \ --basecalled_fastq /fastq_pass/ \ --fast5_pass /fast5_pass/ \ --sequencing_summary sequencing_summary_FAK72834_298b7829.txt \ --outdir nanopore_nanopolishnextflow run main.nf \ -profile singularity \ --nanopolish \ --prefix "test_nanopore_nanopolish" \ --basecalled_fastq /fastq_pass/ \ --fast5_pass /fast5_pass/ \ --sequencing_summary sequencing_summary_FAK72834_298b7829.txt \ --outdir nanopore_nanopolishnextflow run main.nf \ -profile singularity \ --nanopolish \ --prefix "test_nanopore_nanopolish" \ --basecalled_fastq /fastq_pass/ \ --fast5_pass /fast5_pass/ \ --sequencing_summary sequencing_summary_FAK72834_298b7829.txt \ --outdir nanopore_nanopolishnextflow run main.nf \ -profile singularity \ --nanopolish \ --prefix "test_nanopore_nanopolish" \ --basecalled_fastq /fastq_pass/ \ --fast5_pass /fast5_pass/ \ --sequencing_summary sequencing_summary_FAK72834_298b7829.txt \ --outdir nanopore_nanopolishnextflow run main.nf \ -profile singularity \ --nanopolish \ --prefix "test_nanopore_nanopolish" \ --basecalled_fastq /fastq_pass/ \ --fast5_pass /fast5_pass/ \ --sequencing_summary sequencing_summary_FAK72834_298b7829.txt \ --outdir nanopore_nanopolishnextflow run main.nf \ -profile singularity \ --nanopolish \ --prefix "test_nanopore_nanopolish" \ --basecalled_fastq /fastq_pass/ \ --fast5_pass /fast5_pass/ \ --sequencing_summary sequencing_summary_FAK72834_298b7829.txt \ --outdir nanopore_nanopolish
nextflow run main.nf -profile singularity \ --medaka --prefix "test_nanopore_medaka" \ --basecalled_fastq /fastq_pass/ \ --outdir nanopore_medakanextflow run main.nf -profile singularity \ --medaka --prefix "test_nanopore_medaka" \ --basecalled_fastq /fastq_pass/ \ --outdir nanopore_medakanextflow run main.nf -profile singularity \ --medaka --prefix "test_nanopore_medaka" \ --basecalled_fastq /fastq_pass/ \ --outdir nanopore_medaka
Pipeline processes
| Illumina processes | Nanopore medaka processes | Nanopore nanopolish processes |
|---|---|---|
| articDownloadScheme | articDownloadScheme | articDownloadScheme |
| indexReference | - | - |
| versions | versions | versions |
| pangoversions | pangoversions | pangoversions |
| fastqc | fastqcNanopore | fastqcNanopore |
| - | - | pycoqc |
| readTrimming | articRemoveUnmappedReads | articRemoveUnmappedReads |
| - | articGuppyPlex | articGuppyPlex |
| - | articMinIONMedaka | articMinIONNanopolish |
| readMapping | - | - |
| flagStat | - | - |
| trimPrimerSequences | - | - |
| depth | - | - |
| callConsensusFreebayes | - | - |
| annotationVEP | - | - |
| callVariants | - | - |
| makeConsensus | - | - |
| makeQCCSV | makeQCCSV | makeQCCSV |
| writeQCSummaryCSV | writeQCSummaryCSV | writeQCSummaryCSV |
| statsCoverage | - | - |
| statsInsert | - | - |
| statsAlignment | - | - |
| multiqc | multiqcNanopore | multiqcNanopore |
| collateSamples | collateSamples | collateSamples |
| pangolinTyping | pangolinTyping | pangolinTyping |
| nextclade | nextclade | nextclade |
| getVariantDefinitions | getVariantDefinitions | getVariantDefinitions |
| makeReport | makeReport | makeReport |
Let’s have a quick one minute demo
How the results look?
example: Output of Illumina workflow run
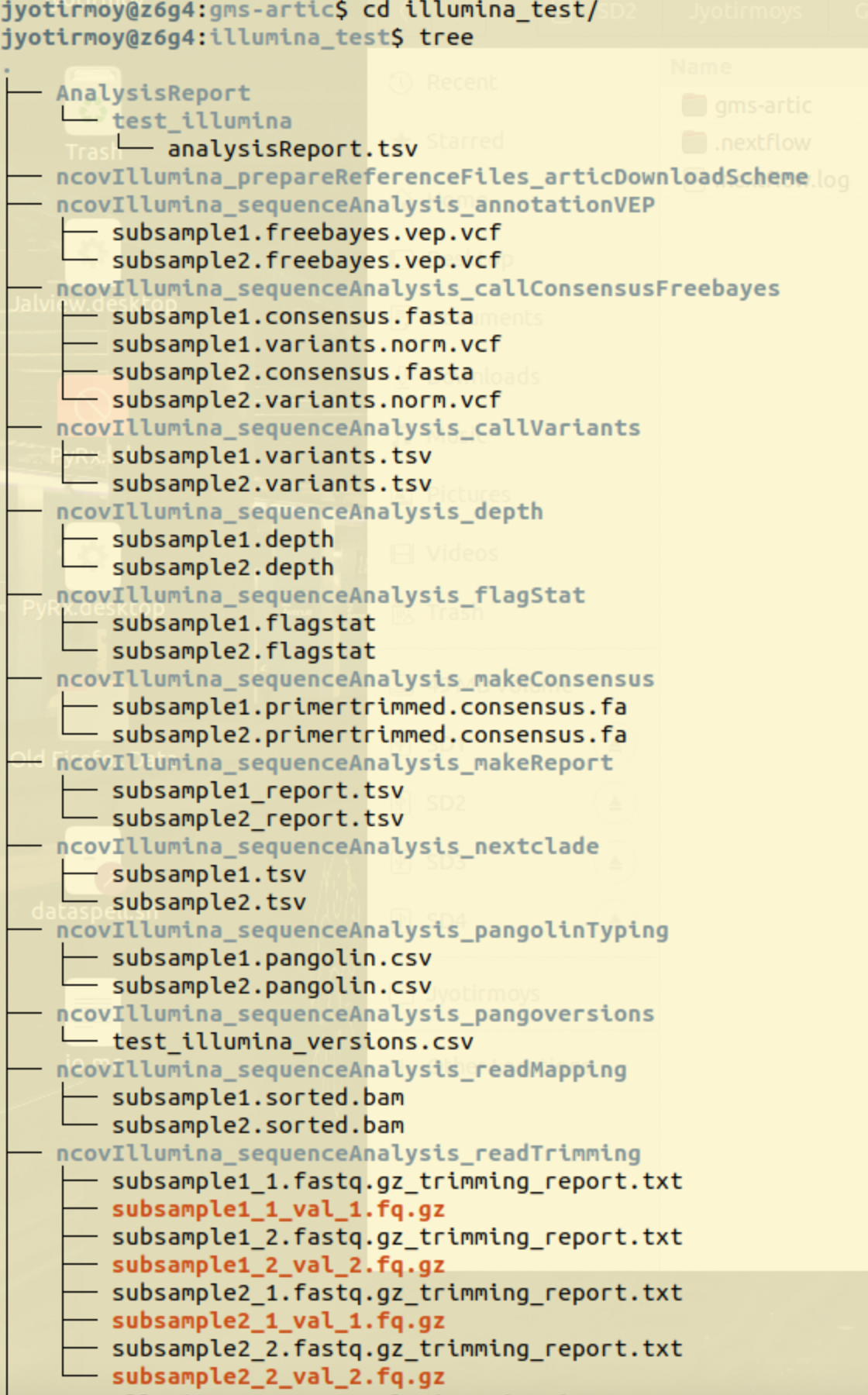
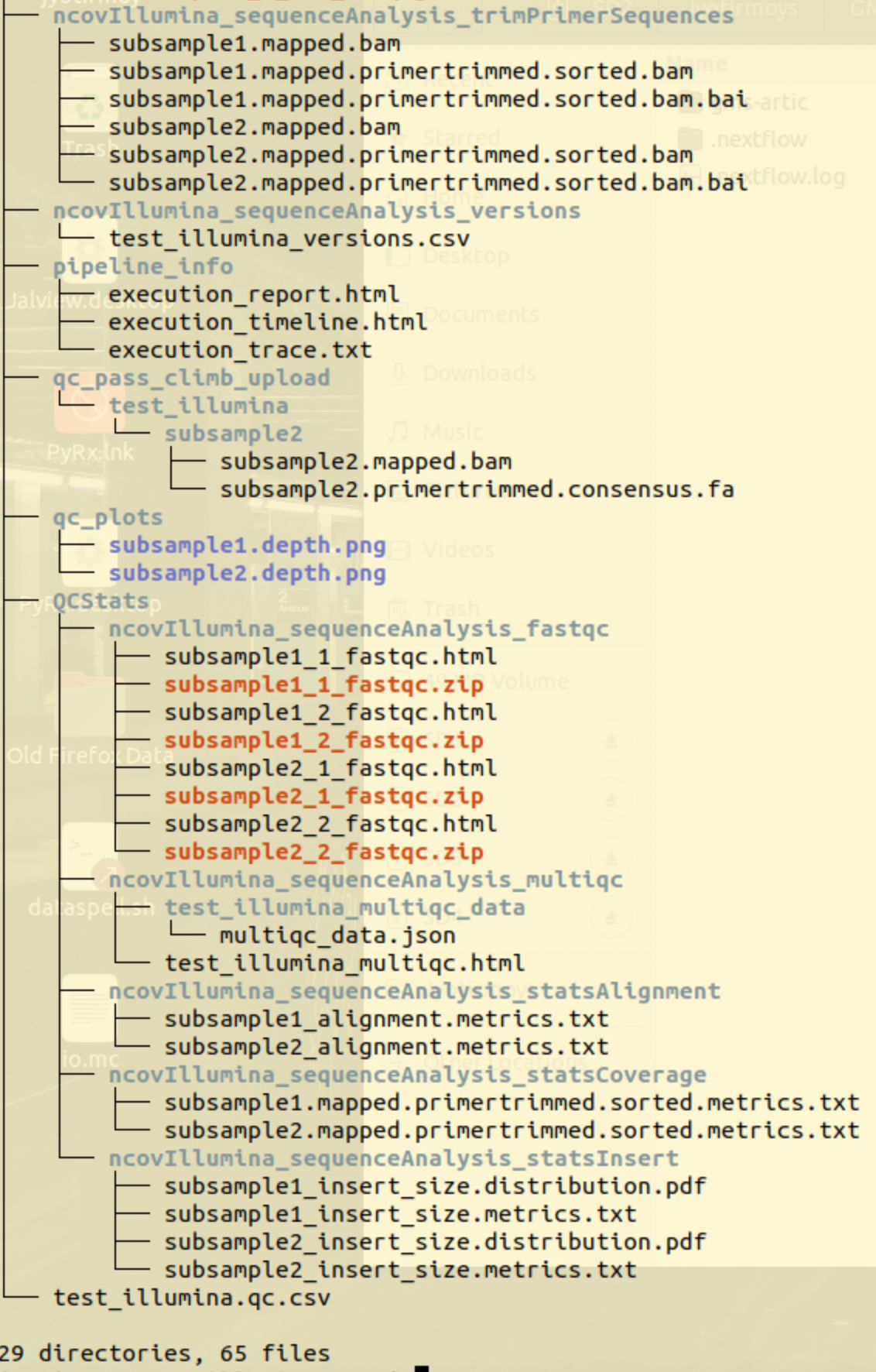
Interactive Result
Where to run?
NGP-server @ Gothenburg University
- SGE cluster
- supports MPI
Local run?
local-server
- Singularity/Conda, Nextflow installation
- AMD64 OS arch (only AMD64 supported container)
Acknowledgements
- Genomic Medicine, Sweden
- Clinical Genomics:
- Gothenburg
- Linköping
- Lund
- Örebro
- Stockholm
- Umeå
- Uppsala
- all USERS of the pipeline!

presenter: Jyotirmoy Das
GMS-Artic A nextflow pipeline with a GMS touch for running the ARTIC network’s fieldbioinformatics tools Jyotirmoy Das Sofia Stamouli Henning Onsbring Tanja Normark Isak Sylvin